| Home | Manual | Algorithm | Contact | Version 1.0 |
| K-mer | P-value ≤ | N | B | n | b | Enrichment |
| UAUAU | 3.4E-254 | 9995 | 1477 | 993 | 578 | 3.94 |
| UAAAU | 3.8E-215 | 9995 | 1829 | 1001 | 601 | 3.28 |
| UGUAUA | 1.1E-209 | 9995 | 702 | 997 | 378 | 5.40 |
| UGUAUAU | 3.3E-206 | 9995 | 435 | 993 | 299 | 6.92 |
| AUAUA | 1.6E-190 | 9995 | 1280 | 991 | 481 | 3.79 |
| UGUAAAU | 1.9E-189 | 9995 | 474 | 1001 | 300 | 6.32 |
| UGUAU | 2.8E-188 | 9995 | 1707 | 999 | 553 | 3.24 |
| UGUAAA | 3.5E-187 | 9995 | 868 | 1001 | 398 | 4.58 |
| UAUAUA | 2.7E-186 | 9995 | 613 | 991 | 335 | 5.51 |
| GUAUAU | 2.7E-183 | 9995 | 572 | 993 | 322 | 5.67 |
| GUAUA | 1.8E-165 | 9995 | 1073 | 997 | 418 | 3.91 |
| UGUAA | 1.7E-163 | 9995 | 1699 | 1002 | 525 | 3.08 |
| UAAAUA | 1.6E-161 | 9995 | 748 | 1000 | 347 | 4.64 |
| GUAAAU | 2.9E-159 | 9995 | 652 | 947 | 315 | 5.10 |
| AAAUA | 4.1E-147 | 9995 | 2024 | 1000 | 553 | 2.73 |
| AUAUU | 2.4E-144 | 9995 | 1746 | 994 | 507 | 2.92 |
| UUGUA | 1.2E-138 | 9995 | 1667 | 1022 | 496 | 2.91 |
| UGUAUAUA | 3.2E-135 | 9995 | 229 | 887 | 173 | 8.51 |
| UAUAUU | 7.3E-131 | 9995 | 673 | 991 | 299 | 4.48 |
| GUAUAUA | 1.1E-130 | 9995 | 265 | 991 | 189 | 7.19 |
| GUAAA | 1.2E-128 | 9995 | 1430 | 1001 | 440 | 3.07 |
| AUAUAU | 9.2E-125 | 9995 | 564 | 991 | 268 | 4.79 |
| UAUAA | 5.2E-124 | 9995 | 1270 | 963 | 399 | 3.26 |
| AUGUA | 3.0E-123 | 9995 | 1512 | 996 | 445 | 2.95 |
| UGUAAAUA | 1.7E-121 | 9995 | 255 | 956 | 177 | 7.26 |
| UAUUU | 8.4E-120 | 9995 | 2333 | 1000 | 559 | 2.39 |
| GUAAAUA | 2.3E-113 | 9995 | 316 | 956 | 190 | 6.29 |
| AAUAU | 3.1E-109 | 9995 | 1580 | 1000 | 438 | 2.77 |
| UAUAUAU | 4.2E-105 | 9995 | 290 | 991 | 178 | 6.19 |
| AUUUU | 1.1E-104 | 9995 | 2760 | 1001 | 591 | 2.14 |
| UUUAU | 2.9E-98 | 9995 | 2108 | 998 | 497 | 2.36 |
| UUAUA | 1.4E-97 | 9995 | 1365 | 998 | 388 | 2.85 |
| AUAUUU | 1.6E-97 | 9995 | 889 | 1000 | 307 | 3.45 |
| AAAUAU | 4.5E-93 | 9995 | 755 | 1000 | 276 | 3.65 |
| UUUGUA | 2.7E-91 | 9995 | 786 | 1022 | 283 | 3.52 |
| AUAAA | 3.3E-91 | 9995 | 1749 | 1000 | 437 | 2.50 |
| UUUUA | 3.6E-86 | 9995 | 2514 | 1000 | 531 | 2.11 |
| UAUUUU | 5.4E-85 | 9995 | 1138 | 1000 | 334 | 2.93 |
| UAAAUAU | 7.1E-85 | 9995 | 308 | 905 | 160 | 5.74 |
| CUGUA | 1.4E-84 | 9995 | 1412 | 1000 | 377 | 2.67 |
| UAAAA | 1.5E-83 | 9995 | 2065 | 1001 | 469 | 2.27 |
| AAUAUA | 2.3E-82 | 9995 | 499 | 905 | 201 | 4.45 |
| UUUAA | 4.2E-82 | 9995 | 2143 | 1001 | 477 | 2.22 |
| AUAUAA | 4.0E-80 | 9995 | 472 | 968 | 199 | 4.35 |
| AAUAA | 8.8E-80 | 9995 | 1535 | 954 | 377 | 2.57 |
| UUUAUA | 1.5E-79 | 9995 | 688 | 1013 | 248 | 3.56 |
| UAUAUUU | 1.1E-78 | 9995 | 323 | 991 | 164 | 5.12 |
| UAUGUA | 6.8E-78 | 9995 | 510 | 991 | 207 | 4.09 |
| UAUAUAA | 7.6E-78 | 9995 | 228 | 954 | 135 | 6.20 |
| UUGUAAA | 10.0E-78 | 9995 | 311 | 1022 | 162 | 5.09 |
| UUGUAA | 2.2E-77 | 9995 | 597 | 1022 | 228 | 3.74 |
| UUAAA | 8.9E-76 | 9995 | 2046 | 1006 | 455 | 2.21 |
| AUGUAU | 1.1E-74 | 9995 | 540 | 1034 | 214 | 3.83 |
| AUAUAUA | 2.2E-73 | 9995 | 217 | 893 | 125 | 6.45 |
| UUAUAU | 8.5E-72 | 9995 | 542 | 985 | 206 | 3.86 |
| UAAAUU | 2.3E-71 | 9995 | 596 | 1000 | 218 | 3.66 |
| AUGUAUA | 2.9E-71 | 9995 | 234 | 991 | 133 | 5.73 |
| UUUUU | 3.6E-71 | 9995 | 2497 | 1006 | 505 | 2.01 |
| UAAUA | 2.6E-70 | 9995 | 1072 | 990 | 300 | 2.83 |
| AUUUUU | 3.6E-69 | 9995 | 1133 | 1006 | 311 | 2.73 |
| UUGUAAAU | 1.0E-68 | 9995 | 167 | 880 | 106 | 7.21 |
| UUGUAUA | 2.1E-67 | 9995 | 225 | 991 | 127 | 5.69 |
| UAUAAA | 3.5E-67 | 9995 | 562 | 962 | 202 | 3.73 |
| AUAUGUA | 8.1E-67 | 9995 | 218 | 899 | 120 | 6.12 |
| AAAUU | 2.4E-66 | 9995 | 1845 | 1001 | 410 | 2.22 |
| UUGUAU | 8.4E-66 | 9995 | 598 | 1006 | 212 | 3.52 |
| AAUAUAU | 1.4E-65 | 9995 | 221 | 986 | 124 | 5.69 |
| UAUGU | 1.5E-65 | 9995 | 1365 | 986 | 337 | 2.50 |
| UAAAAU | 1.8E-65 | 9995 | 735 | 994 | 236 | 3.23 |
| ACUGUA | 2.9E-65 | 9995 | 483 | 996 | 187 | 3.89 |
| AAAAU | 2.3E-64 | 9995 | 2276 | 1001 | 463 | 2.03 |
| AUAUGU | 2.3E-64 | 9995 | 468 | 899 | 174 | 4.13 |
| AUAAU | 3.2E-64 | 9995 | 1129 | 986 | 299 | 2.68 |
| AAAUAA | 1.3E-62 | 9995 | 771 | 953 | 233 | 3.17 |
| UUGUAUAU | 1.8E-62 | 9995 | 140 | 886 | 93 | 7.49 |
| AUUAU | 2.2E-62 | 9995 | 1239 | 981 | 312 | 2.57 |
| AUGUAUAU | 2.9E-62 | 9995 | 145 | 991 | 98 | 6.82 |
| UGUAUAUU | 2.2E-61 | 9995 | 129 | 965 | 91 | 7.31 |
| UUUUAA | 9.2E-61 | 9995 | 1091 | 1000 | 290 | 2.66 |
| AUUGUA | 1.0E-60 | 9995 | 438 | 1020 | 174 | 3.89 |
| GUAUAUAU | 8.2E-60 | 9995 | 119 | 991 | 87 | 7.37 |
| UGUAUAUAU | 1.1E-59 | 9995 | 105 | 887 | 79 | 8.48 |
| UUUGU | 1.2E-59 | 9995 | 2181 | 1024 | 449 | 2.01 |
| UGUAAAUU | 3.6E-59 | 9995 | 130 | 908 | 88 | 7.45 |
| UGUAAAUAU | 1.3E-58 | 9995 | 103 | 905 | 78 | 8.36 |
| UUUAAA | 1.4E-58 | 9995 | 1013 | 1006 | 275 | 2.70 |
| UAAAUAA | 1.4E-58 | 9995 | 254 | 999 | 127 | 5.00 |
| UUUUUA | 3.8E-58 | 9995 | 1062 | 999 | 281 | 2.65 |
| UGUACA | 5.4E-58 | 9995 | 513 | 1038 | 188 | 3.53 |
| AUAUAUU | 5.5E-58 | 9995 | 254 | 991 | 126 | 5.00 |
| AAUUU | 6.8E-58 | 9995 | 1913 | 1006 | 405 | 2.10 |
| UAUAUAUA | 4.6E-57 | 9995 | 124 | 893 | 84 | 7.58 |
| AUUUA | 5.7E-57 | 9995 | 1623 | 995 | 361 | 2.23 |
| UAUUUA | 1.9E-56 | 9995 | 609 | 972 | 198 | 3.34 |
| AUGUAAAU | 2.5E-56 | 9995 | 149 | 1001 | 95 | 6.37 |
| UAUAC | 3.9E-56 | 9995 | 757 | 1013 | 229 | 2.98 |
| AAUAAA | 3.9E-56 | 9995 | 802 | 984 | 233 | 2.95 |
| UAUUA | 9.5E-56 | 9995 | 1108 | 986 | 282 | 2.58 |
| CUGUAUAU | 2.2E-55 | 9995 | 98 | 905 | 74 | 8.34 |
| AUAAAU | 2.4E-55 | 9995 | 570 | 999 | 192 | 3.37 |
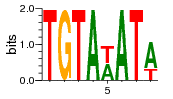
| Motif 1: | |
|
P-value ≤ 4.9E-324
There are B=702 occurrences of the motif, out of which b=422 occur at the top n=789 sequences.
|
|
Download logo in
format
|
Clicking this button opens a pop-up window presenting the occurrences of the motif in the input sequences, aligned to each other.
The occurrences are colored by the logo color scheme and the flanking positions are colored in black.
Each occurrence is identified by the title of the sequence in which it occurs, the index of the sequence in the query list, the starting position of the occurrence in the sequence and the strand (in double-strand search mode only).
Clicking this button opens a pop-up window showing a schematic presentation of the distribution of the motif occurrences in the query sequences.
Each line represents an input sequence (following the original order), where a bold line is a sequence containing the motif.
The occurrences are represented by colored blocks (fuchsia for the occurrences at the plus strand and blue for the occurrences at the minus strand).
Placing the cursor on a motif occurrence reveals the title of the sequence in which it occurs, the starting position of the occurrence in the sequence and the strand (in double-strand search mode only).
|

 View list
View list